Calculate a suite of multispectral indices such as NDVI, SAVI etc. in an efficient way via C++.
Usage
spectralIndices(
img,
blue = NULL,
green = NULL,
red = NULL,
nir = NULL,
redEdge1 = NULL,
redEdge2 = NULL,
redEdge3 = NULL,
swir1 = NULL,
swir2 = NULL,
swir3 = NULL,
scaleFactor = 1,
skipRefCheck = FALSE,
indices = NULL,
index = NULL,
maskLayer = NULL,
maskValue = 1,
coefs = list(L = 0.5, G = 2.5, L_evi = 1, C1 = 6, C2 = 7.5, s = 1, swir2ccc = NULL,
swir2coc = NULL),
...
)Arguments
- img
SpatRaster. Typically remote sensing imagery, which is to be classified.
- blue
Character or integer. Blue band.
- green
Character or integer. Green band.
- red
Character or integer. Red band.
- nir
Character or integer. Near-infrared band (700-1100nm).
- redEdge1
Character or integer. Red-edge band (705nm)
- redEdge2
Character or integer. Red-edge band (740nm)
- redEdge3
Character or integer. Red-edge band (783nm)
- swir1
not used
- swir2
Character or integer. Short-wave-infrared band (1400-1800nm).
- swir3
Character or integer. Short-wave-infrared band (2000-2500nm).
- scaleFactor
Numeric. Scale factor for the conversion of scaled reflectances to [0,1] value range (applied as reflectance/scaleFactor) Neccesary for calculating EVI/EVI2 with scaled reflectance values.
- skipRefCheck
Logical. When EVI/EVI2 is to be calculated there is a rough heuristic check, whether the data are inside [0,1]+/-0.5 (after applying a potential
scaleFactor). If there are invalid reflectances, e.g. clouds with reflectance > 1 this check will result in a false positive and skip EVI calculation. Use this argument to skip this check in such cases *iff* you are sure the data and scaleFactor are valid.- indices
Character. One or more spectral indices to calculate (see Details). By default (NULL) all implemented indices given the spectral bands which are provided will be calculated.
- index
Character. Alias for
indices.- maskLayer
RasterLayer or SpatRaster containing a mask, e.g. clouds, for which pixels are set to NA. Alternatively a layername or -number can be provided if the mask is part of
img.- maskValue
Integer. Pixel value in
maskLayerwhich should be masked in output, i.e. will be set toNAin all calculated indices.- coefs
List of coefficients (see Details).
- ...
further arguments such as filename etc. passed to writeRaster
Details
spectralIndices calculates all indices in one go in C++, which is more efficient than calculating each index separately (for large rasters).
By default all indices which can be calculated given the specified indices will be calculated. If you don't want all indices, use the indices argument to specify exactly which indices are to be calculated.
See the table bellow for index names and required bands.
Index values outside the valid value ranges (if such a range exists) will be set to NA. For example a pixel with NDVI > 1 will be set to NA.
| Index | Description | Source | Bands | Formula |
| CLG | Green-band Chlorophyll Index | Gitelson2003 | redEdge3, green | \(redEdge3/green - 1\) |
| CLRE | Red-edge-band Chlorophyll Index | Gitelson2003 | redEdge3, redEdge1 | \(redEdge3/redEdge1 - 1\) |
| CTVI | Corrected Transformed Vegetation Index | Perry1984 | red, nir | \((NDVI + 0.5)/sqrt(abs(NDVI + 0.5))\) |
| DVI | Difference Vegetation Index | Richardson1977 | red, nir | \(s * nir - red\) |
| EVI | Enhanced Vegetation Index | Huete1999 | red, nir, blue | \(G * ((nir - red)/(nir + C1 * red - C2 * blue + L_evi))\) |
| EVI2 | Two-band Enhanced Vegetation Index | Jiang 2008 | red, nir | \(G * (nir - red)/(nir + 2.4 * red + 1)\) |
| GEMI | Global Environmental Monitoring Index | Pinty1992 | red, nir | \((((nir^2 - red^2) * 2 + (nir * 1.5) + (red * 0.5))/(nir + red + 0.5)) * (1 - ((((nir^2 - red^2) * 2 + (nir * 1.5) + (red * 0.5))/(nir + red + 0.5)) * 0.25)) - ((red - 0.125)/(1 - red))\) |
| GNDVI | Green Normalised Difference Vegetation Index | Gitelson1998 | green, nir | \((nir - green)/(nir + green)\) |
| KNDVI | Kernel Normalised Difference Vegetation Index | Camps-Valls2021 | red, nir | \(tanh(((nir - red)/(nir + red)))^2\) |
| MCARI | Modified Chlorophyll Absorption Ratio Index | Daughtery2000 | green, red, redEdge1 | \(((redEdge1 - red) - (redEdge1 - green)) * (redEdge1/red)\) |
| MNDWI | Modified Normalised Difference Water Index | Xu2006 | green, swir2 | \((green - swir2)/(green + swir2)\) |
| MSAVI | Modified Soil Adjusted Vegetation Index | Qi1994 | red, nir | \(nir + 0.5 - (0.5 * sqrt((2 * nir + 1)^2 - 8 * (nir - (2 * red))))\) |
| MSAVI2 | Modified Soil Adjusted Vegetation Index 2 | Qi1994 | red, nir | \((2 * (nir + 1) - sqrt((2 * nir + 1)^2 - 8 * (nir - red)))/2\) |
| MTCI | MERIS Terrestrial Chlorophyll Index | DashAndCurran2004 | red, redEdge1, redEdge2 | \((redEdge2 - redEdge1)/(redEdge1 - red)\) |
| NBRI | Normalised Burn Ratio Index | Garcia1991 | nir, swir3 | \((nir - swir3)/(nir + swir3)\) |
| NDREI1 | Normalised Difference Red Edge Index 1 | GitelsonAndMerzlyak1994 | redEdge2, redEdge1 | \((redEdge2 - redEdge1)/(redEdge2 + redEdge1)\) |
| NDREI2 | Normalised Difference Red Edge Index 2 | Barnes2000 | redEdge3, redEdge1 | \((redEdge3 - redEdge1)/(redEdge3 + redEdge1)\) |
| NDVI | Normalised Difference Vegetation Index | Rouse1974 | red, nir | \((nir - red)/(nir + red)\) |
| NDVIC | Corrected Normalised Difference Vegetation Index | Nemani1993 | red, nir, swir2 | \((nir - red)/(nir + red) * (1 - ((swir2 - swir2ccc)/(swir2coc - swir2ccc)))\) |
| NDWI | Normalised Difference Water Index | McFeeters1996 | green, nir | \((green - nir)/(green + nir)\) |
| NDWI2 | Normalised Difference Water Index | Gao1996 | nir, swir2 | \((nir - swir2)/(nir + swir2)\) |
| NRVI | Normalised Ratio Vegetation Index | Baret1991 | red, nir | \((red/nir - 1)/(red/nir + 1)\) |
| REIP | Red Edge Inflection Point | GuyotAndBarnet1988 | red, redEdge1, redEdge2, redEdge3 | \(0.705 + 0.35 * ((red + redEdge3)/(2 - redEdge1))/(redEdge2 - redEdge1)\) |
| RVI | Ratio Vegetation Index | red, nir | \(red/nir\) | |
| SATVI | Soil Adjusted Total Vegetation Index | Marsett2006 | red, swir2, swir3 | \((swir2 - red)/(swir2 + red + L) * (1 + L) - (swir3/2)\) |
| SAVI | Soil Adjusted Vegetation Index | Huete1988 | red, nir | \((nir - red) * (1 + L)/(nir + red + L)\) |
| SLAVI | Specific Leaf Area Vegetation Index | Lymburger2000 | red, nir, swir2 | \(nir/(red + swir2)\) |
| SR | Simple Ratio Vegetation Index | Birth1968 | red, nir | \(nir/red\) |
| TTVI | Thiam's Transformed Vegetation Index | Thiam1997 | red, nir | \(sqrt(abs((nir - red)/(nir + red) + 0.5))\) |
| TVI | Transformed Vegetation Index | Deering1975 | red, nir | \(sqrt((nir - red)/(nir + red) + 0.5)\) |
| WDVI | Weighted Difference Vegetation Index | Richardson1977 | red, nir | \(nir - s * red\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
| CUSTOM | Super custom index | Mueller2024 | red | \(red * 0\) |
| CUSTOM2 | Super custom index | Mueller2024 | swir1 | \(swir1 - swir1\) |
Some indices require additional parameters, such as the slope of the soil line which are specified via a list to the coefs argument.
Although the defaults are sensible values, values like the soil brightnes factor L for SAVI should be adapted depending on the characteristics of the scene.
The coefficients are:
| Coefficient | Description | Affected Indices |
s | slope of the soil line | DVI, WDVI |
L_evi, C1, C2, G | various | EVI |
L | soil brightness factor | SAVI, SATVI |
swir2ccc | minimum swir2 value (completely closed forest canopy) | NDVIC |
swir2coc | maximum swir2 value (completely open canopy) | NDVIC |
The wavelength band names are defined following Schowengertd 2007, p10. The last column shows exemplarily which Landsat 5 TM bands correspond to which wavelength range definition.
| Band | Description | Wavl_min | Wavl_max | Landsat5_Band | Sentinel2_Band | vis |
| visible | 400 | 680 | 1,2,3 | 2,3,4 | red-edge1 | red-edge1 |
| 680 | 720 | - | 5 | red-edge2 | red-edge2 | 720 |
| 760 | - | 6 | red-edge3 | red-edge3 | 760 | 800 |
| - | 7 | nir | near infra-red | 800 | 1100 | 4 |
| 8/8a | swir1 | short-wave infra-red | 1100 | 1351 | - | 9,10 |
| swir2 | short-wave infra-red | 1400 | 1800 | 5 | 11 | swir3 |
| short-wave infra-red | 2000 | 2500 | 7 | 12 | mir1 | mid-wave infra-red |
| 3000 | 4000 | - | - | mir2 | mid-wave infra-red | 45000 |
| 5000 | - | - | tir1 | thermal infra-red | 8000 | 9500 |
| - | - | tir2 | thermal infra-red | 10000 | 140000 | 6 |
Examples
library(ggplot2)
library(terra)
## Calculate NDVI
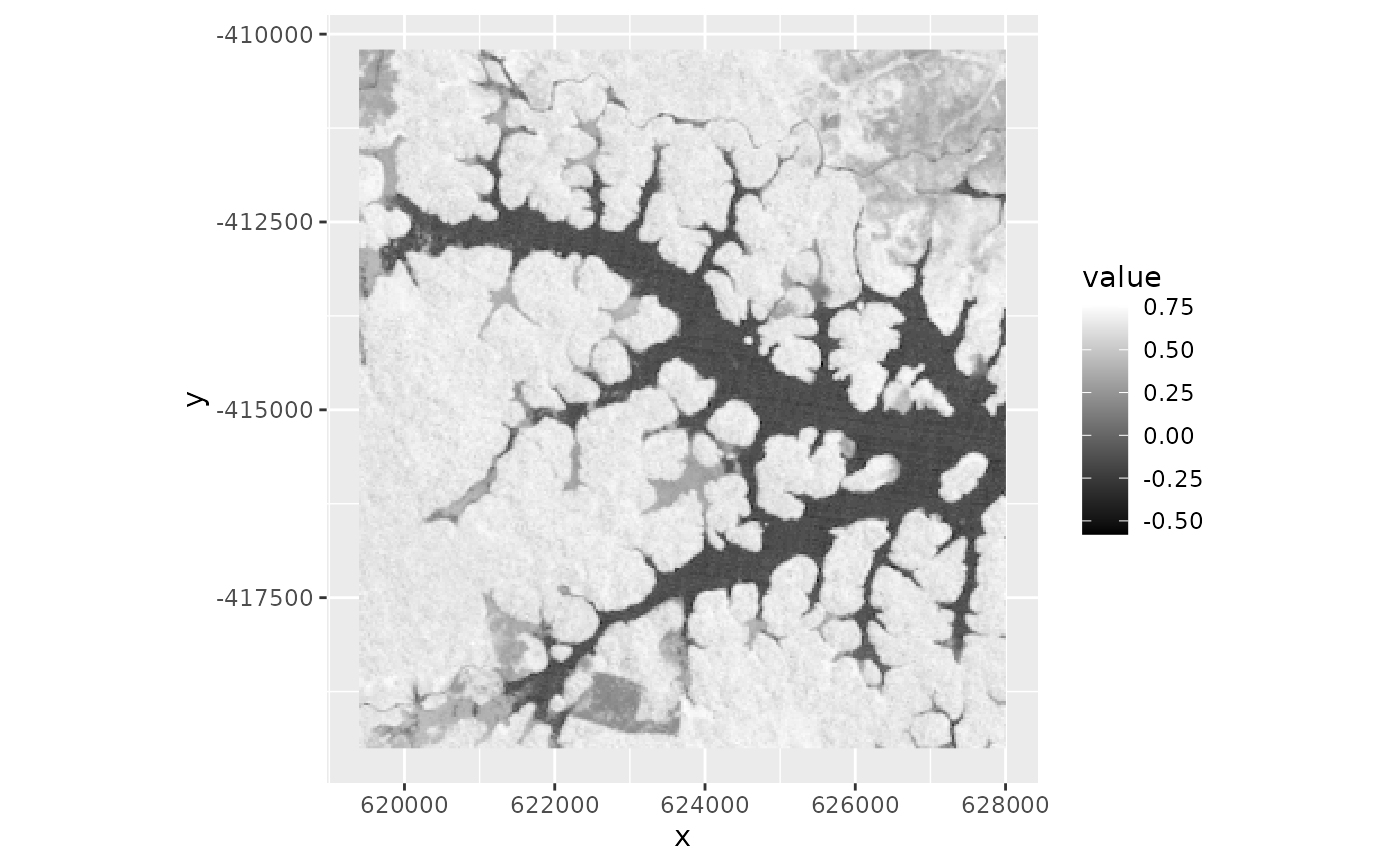
ndvi <- spectralIndices(lsat, red = "B3_dn", nir = "B4_dn", indices = "NDVI")
ndvi
#> class : SpatRaster
#> size : 310, 287, 1 (nrow, ncol, nlyr)
#> resolution : 30, 30 (x, y)
#> extent : 619395, 628005, -419505, -410205 (xmin, xmax, ymin, ymax)
#> coord. ref. : +proj=utm +zone=22 +ellps=WGS84 +towgs84=0,0,0,0,0,0,0 +units=m +no_defs
#> source(s) : memory
#> name : NDVI
#> min value : -0.5789474
#> max value : 0.7629630
ggR(ndvi, geom_raster = TRUE) +
scale_fill_gradientn(colours = c("black", "white"))
 # \donttest{
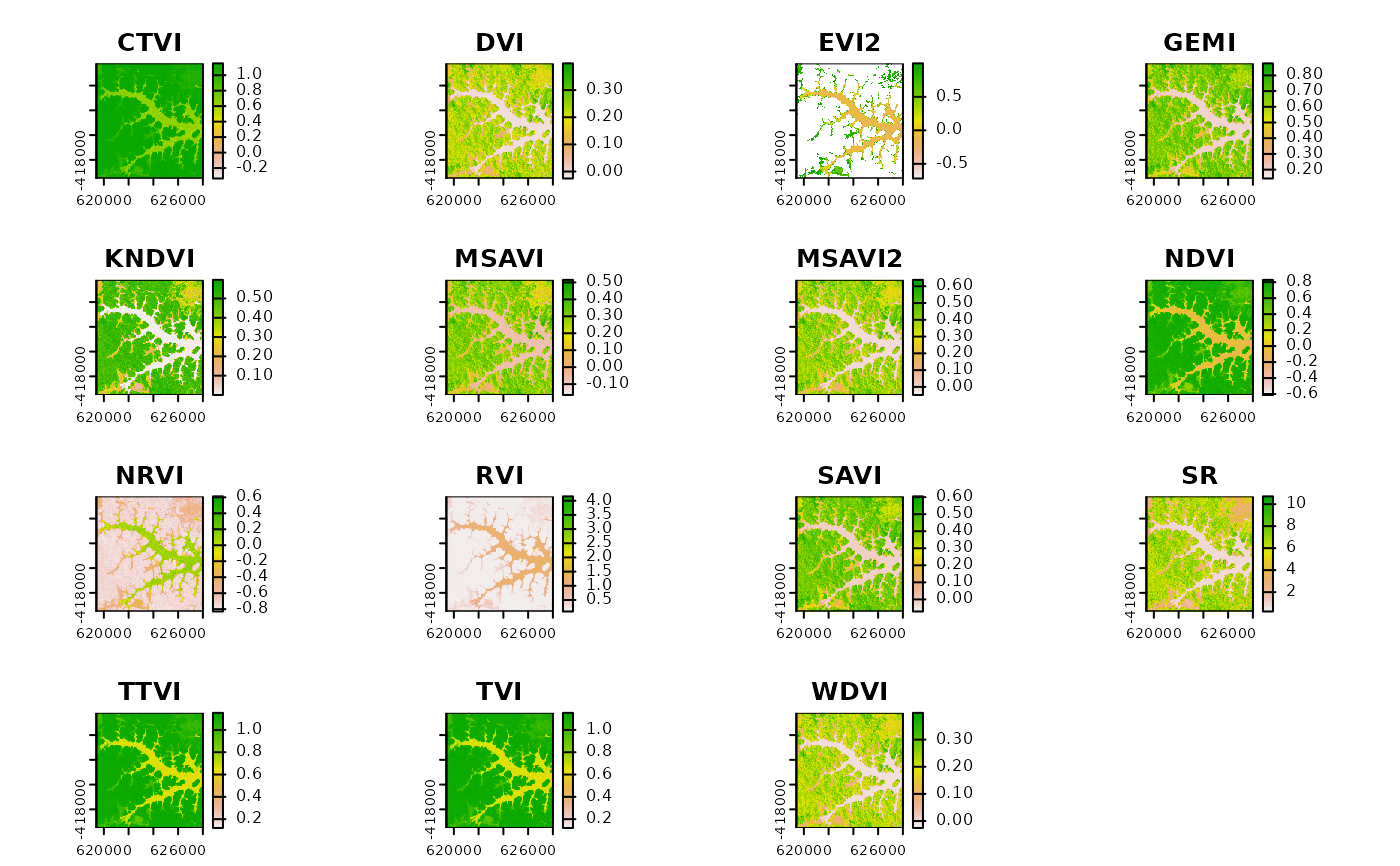
## Calculate all possible indices, given the provided bands
## Convert DNs to reflectance (required to calculate EVI and EVI2)
mtlFile <- system.file("external/landsat/LT52240631988227CUB02_MTL.txt", package="RStoolbox")
lsat_ref <- radCor(lsat, mtlFile, method = "apref")
#> 09:46:54 | Bands to convert to reflectance: B1_dn, B2_dn, B3_dn, B4_dn, B5_dn, B7_dn
#> 09:46:54 | Thermal bands to convert to brightness temperature: B6_dn
#> 09:46:54 | Processing thermal band(s)
#> 09:46:54 | Processing radiance / reflectance
SI <- spectralIndices(lsat_ref, red = "B3_tre", nir = "B4_tre")
plot(SI)
# \donttest{
## Calculate all possible indices, given the provided bands
## Convert DNs to reflectance (required to calculate EVI and EVI2)
mtlFile <- system.file("external/landsat/LT52240631988227CUB02_MTL.txt", package="RStoolbox")
lsat_ref <- radCor(lsat, mtlFile, method = "apref")
#> 09:46:54 | Bands to convert to reflectance: B1_dn, B2_dn, B3_dn, B4_dn, B5_dn, B7_dn
#> 09:46:54 | Thermal bands to convert to brightness temperature: B6_dn
#> 09:46:54 | Processing thermal band(s)
#> 09:46:54 | Processing radiance / reflectance
SI <- spectralIndices(lsat_ref, red = "B3_tre", nir = "B4_tre")
plot(SI)
 ## Custom Spectral Index Calculation (beta) (supports only bands right now...)
# Get all indices
# Supports: Parentheses (), Addition +, Subtraction -, Multiplication *, Division /
idxdb <- getOption("RStoolbox.idxdb")
# Customize the RStoolbox index-database and overwrite the option
cdb <- c(idxdb, CUSTOM = list( list(c("Mueller2024", "Super custom index"),
function(blue, red) {blue + red})))
rsOpts(idxdb = cdb)
# Calculate the custom index, (also together with the provided ones)
custom_ind <- spectralIndices(lsat, blue = 1, red = 3, nir = 4, indices = c("NDVI", "CUSTOM"))
# }
## Custom Spectral Index Calculation (beta) (supports only bands right now...)
# Get all indices
# Supports: Parentheses (), Addition +, Subtraction -, Multiplication *, Division /
idxdb <- getOption("RStoolbox.idxdb")
# Customize the RStoolbox index-database and overwrite the option
cdb <- c(idxdb, CUSTOM = list( list(c("Mueller2024", "Super custom index"),
function(blue, red) {blue + red})))
rsOpts(idxdb = cdb)
# Calculate the custom index, (also together with the provided ones)
custom_ind <- spectralIndices(lsat, blue = 1, red = 3, nir = 4, indices = c("NDVI", "CUSTOM"))
# }
